Recent findings from icddr,b genomics laboratory, published in the prominent Scientific Reports journal (part of the Nature Publishing Group), shed new light on the natural diversity and evolution of CRISPR/Cas system in the bacteriophage viruses that target Vibrio cholerae, the causative agent of cholera.
What is the CRISPR/Cas system?
The CRISPR/Cas system has featured prominently in the news and promises to revolutionise biotechnology and gene editing, and it is worth briefly discussing the history of its discovery to fully understand the significance of these results.
CRISPR-Cas9 Editing of the Genome. Photo: National Human Genome Research Institute. CC BY-NC 2.0
CRISPR stands for Clustered Regularly Interspaced Palindromic Repeats – it is a family of short, repetitive DNA sequences usually found in the genomes of bacteria. Although they were discovered first discovered in 1987, it is only recently that the full significance of these repetitive DNA elements has been fully understood.
Experiments in the early 2000s would show that the repetitive CRISPR sequences are transcribed to RNA, and associated with a specific set of protein-encoding genes. These protein-encoding genes were named CRISPR-associated systems, or cas genes. The combination of these repetitive DNA elements with these protein-encoding genes was termed the CRISP-Cas system.
Further work began to elucidate the function of this CRISPR-Cas system in bacteria. It was found that some of these CRISPR “spacer” elements match genetic sequences found in bacteriophages – “bacteria-eating” viruses that can attack bacterial cells. This suggested that the CRISPR-Cas system could act as a kind of adaptive bacterial immune system, with the CRISPR repeats guiding the Cas proteins to specific genetic sequences in invading viruses.
A major breakthrough came in 2012, through the work of Jennifer Doudna and Emmanuelle Charpentier. They engineered a simplified bacterial CRISPR-Cas system, and showed that it could find and cut the genetic sequences that were specified by the CRISPR RNAs. By altering the sequence of these “guide” RNAs, the system could be engineered to find and cut any arbitrary sequence of DNA.
This ability of the CRISPR/Cas system to find and cut any desired DNA sequence promises to revolutionise gene modification, with enormous potential for biotechnology, medicine and agriculture.
Natural diversity of CRISPR/Cas in bacteriophages in Bangladesh
Although much of the press attention on CRISPR/Cas has focused on its potential for biotechnology, there is still much work to be done on understanding the role and function of the CRISP/Cas system in nature.
It is important to emphasise that CRISPR/Cas was not something made entirely by genetic engineers, but a naturally occurring system which has been repurposed for biotechnology. As a result, it is important to fully understand the scope of its functional diversity to make best use of it for industry and medicine – just as understanding the full diversity of natural antibiotics is important to tackle problems like antibiotic resistance.
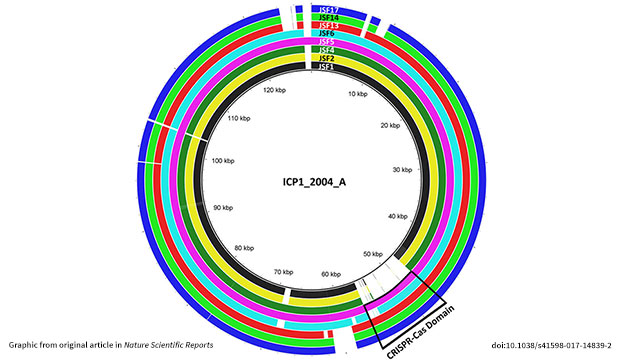
This is why the recent icddr,b findings from a team led by former senior scientist Dr Shah M Faruque are so interesting – they reveal new features of this system in the natural context of Bangladesh. Using DNA sequencing, the paper analyses the CRISPR-Cas system in the bacteriophage viruses that target V. cholerae, the causative agent of cholera.
This system is remarkably different from most known CRISPR-Cas systems in that it is not found in bacteria, but in the viruses that attack bacteria. Normally bacteria use these elements to protect against invading viruses, but in this case viruses are using the system to evade bacterial immune responses. The presence of viral CRISPR/Cas elements in cholera-killing bacteriophages was first described by Andrew Camilli’s group in America, in a paper published in Nature in 2013.
The icddr,b genomics lab team has now built upon that groundbreaking work by analysing phages and V. cholerae strains collected over the multiple cholera outbreaks in Bangladesh. Some were isolated from environmental sources like surface water in Dhaka city, and others were collected from the stools of cholera patients.
Due to the recurring seasonal outbreaks of cholera in Bangladesh, these samples represent a powerful natural experiment to analyse how the CRISPR/Cas system is evolving in these viruses over time. Using DNA sequencing, the authors can locate specific CRISPR spacer sequences in the virus that target the immune system of V.cholerae, allowing the viruses to evade the immune response and infect the bacteria more effectively.
The number of immune-evading CRISPR spacer sequences has progressively increased in viruses isolated over different years. Viruses that have an increased number of these immune-evading spacers were also shown to be more effective in infecting cholera bacteria, suggesting that acquiring more than one CRISPR spacer sequence increases the ability of the viruses to evade the bacterial immune response.
From a scientific perspective, these results are fascinating because they reveal an ongoing evolutionary arms race between V.cholerae and its associated viruses. Previous work from icddr,b genomics lab has shown that these viruses play a critical role in limiting seasonal outbreaks. This work reveals some of the details of the ongoing battle between virus and bacteria on a molecular level.
From a practical standpoint, the work could help in efforts to use these viruses for environmental control of cholera outbreaks, or for direct phage therapy in cholera patients. Viruses could be engineered to have the optimal CRISPR/Cas system to fully evade bacterial immune systems, thus killing them more effectively.
Ultimately, this work demonstrates the value of conducting basic research in developing country contexts and the importance of institutions like icddr,b to support this kind of work. Thanks to this institutional support, the icddr,b genomics lab team was able to combine cutting-edge technology with an intimate understanding of the local ecological and socio-economic setting of Bangladesh. These results contribute to the body of knowledge around one of the most exciting topics in contemporary biology, and could one day help to improve lives not just in Bangladesh but across the entire developing world.

